
cgbind: A Python Module and Web App for Automated Metallocage Construction and Host–Guest Characterization | Journal of Chemical Information and Modeling

Simulated spectrum of isotopic distribution of the peptide sequence... | Download Scientific Diagram
Simplifying MS1 and MS2 spectra to achieve lower mass error, more dynamic range, and higher peptide identification confidence on the Bruker timsTOF Pro | PLOS ONE
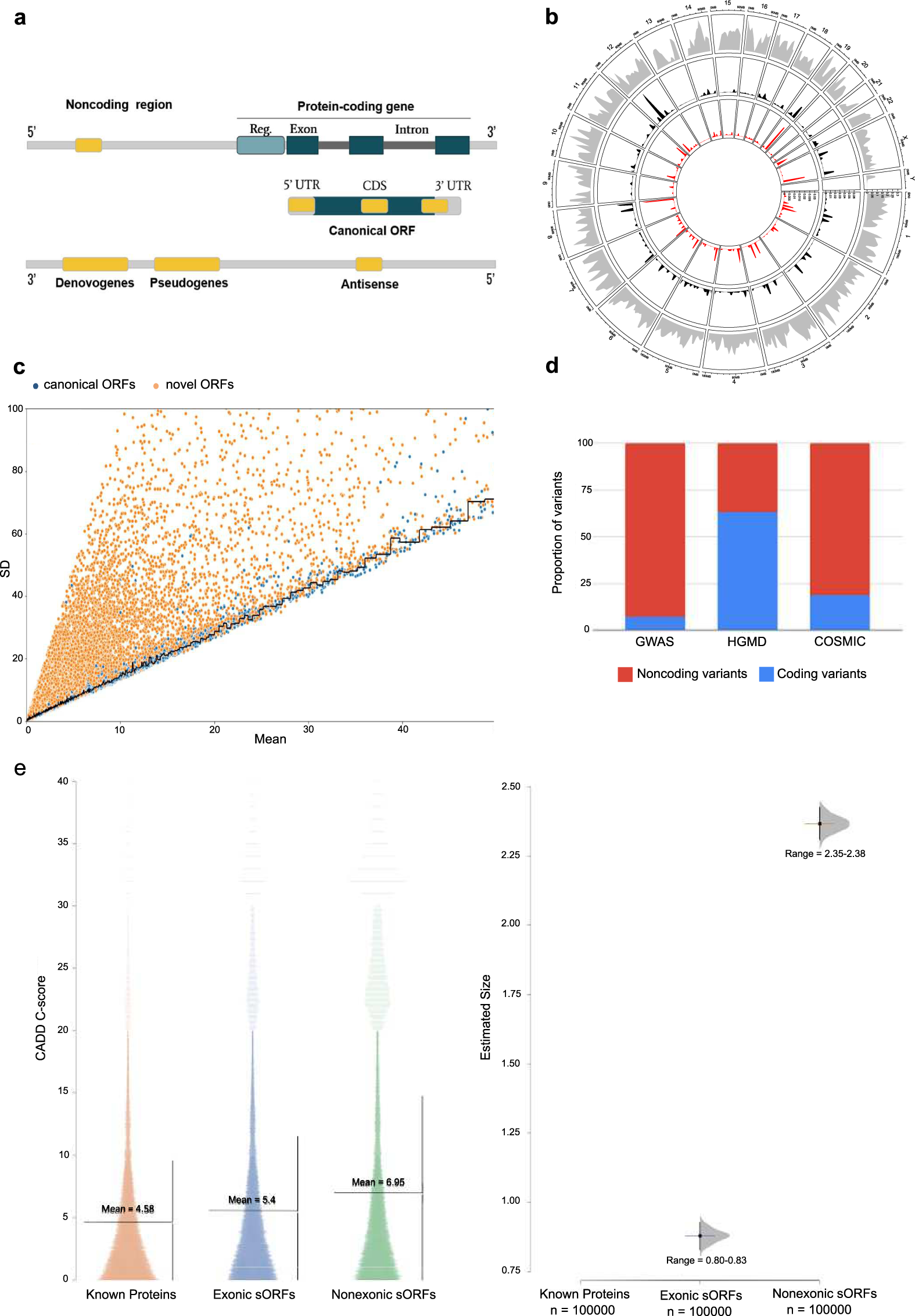
Pan-cancer analysis of transcripts encoding novel open-reading frames (nORFs) and their potential biological functions | npj Genomic Medicine
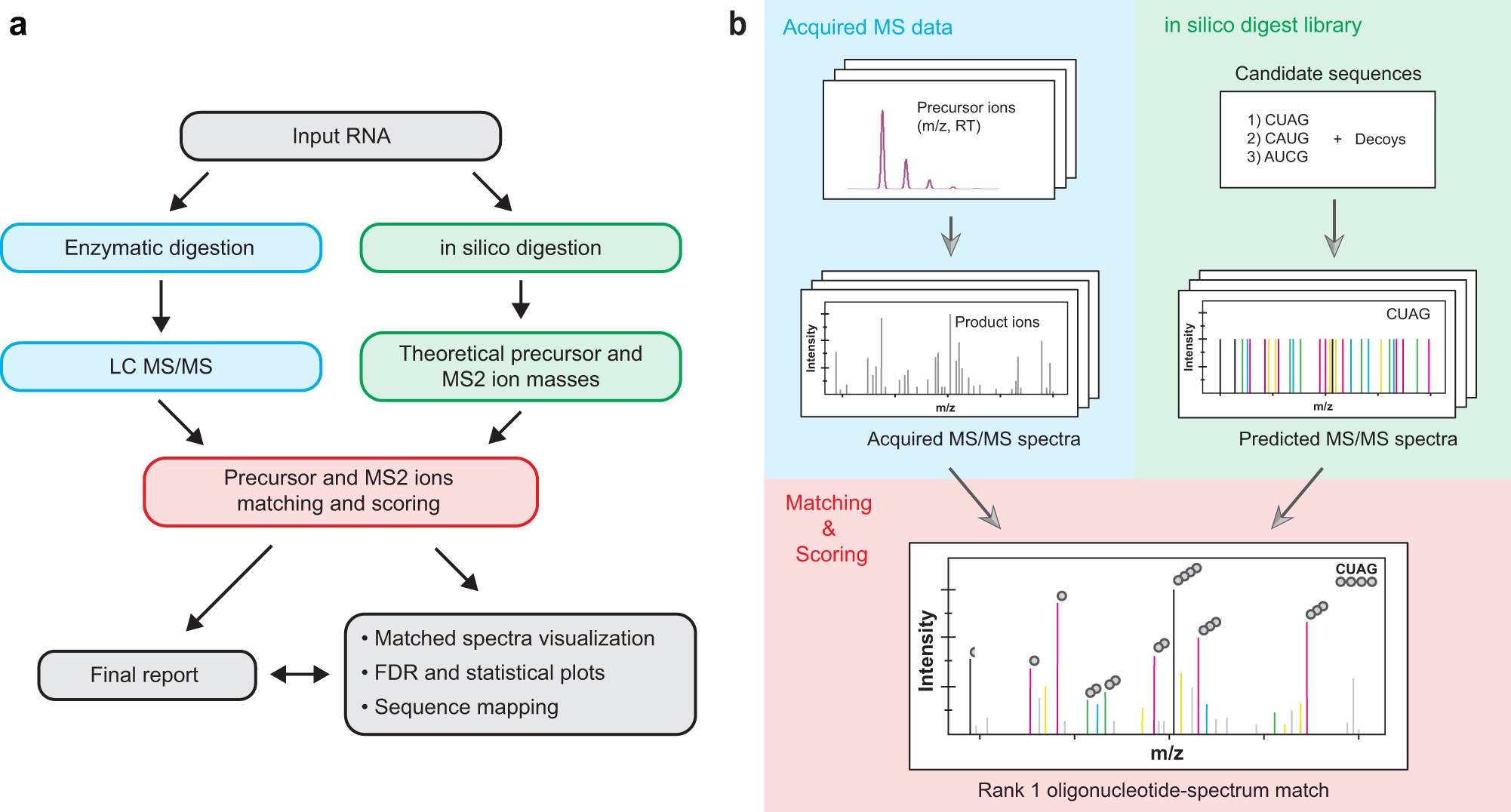
Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry | Nature Communications

Ad hoc learning of peptide fragmentation from mass spectra enables an interpretable detection of phosphorylated and cross-linked peptides | Nature Machine Intelligence

Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1 - ScienceDirect

Ursgal, Universal Python Module Combining Common Bottom-Up Proteomics Tools for Large-Scale Analysis | Journal of Proteome Research

LC–MS peak assignment based on unanimous selection by six machine learning algorithms | Scientific Reports

pytc: Open-Source Python Software for Global Analyses of Isothermal Titration Calorimetry Data | Biochemistry






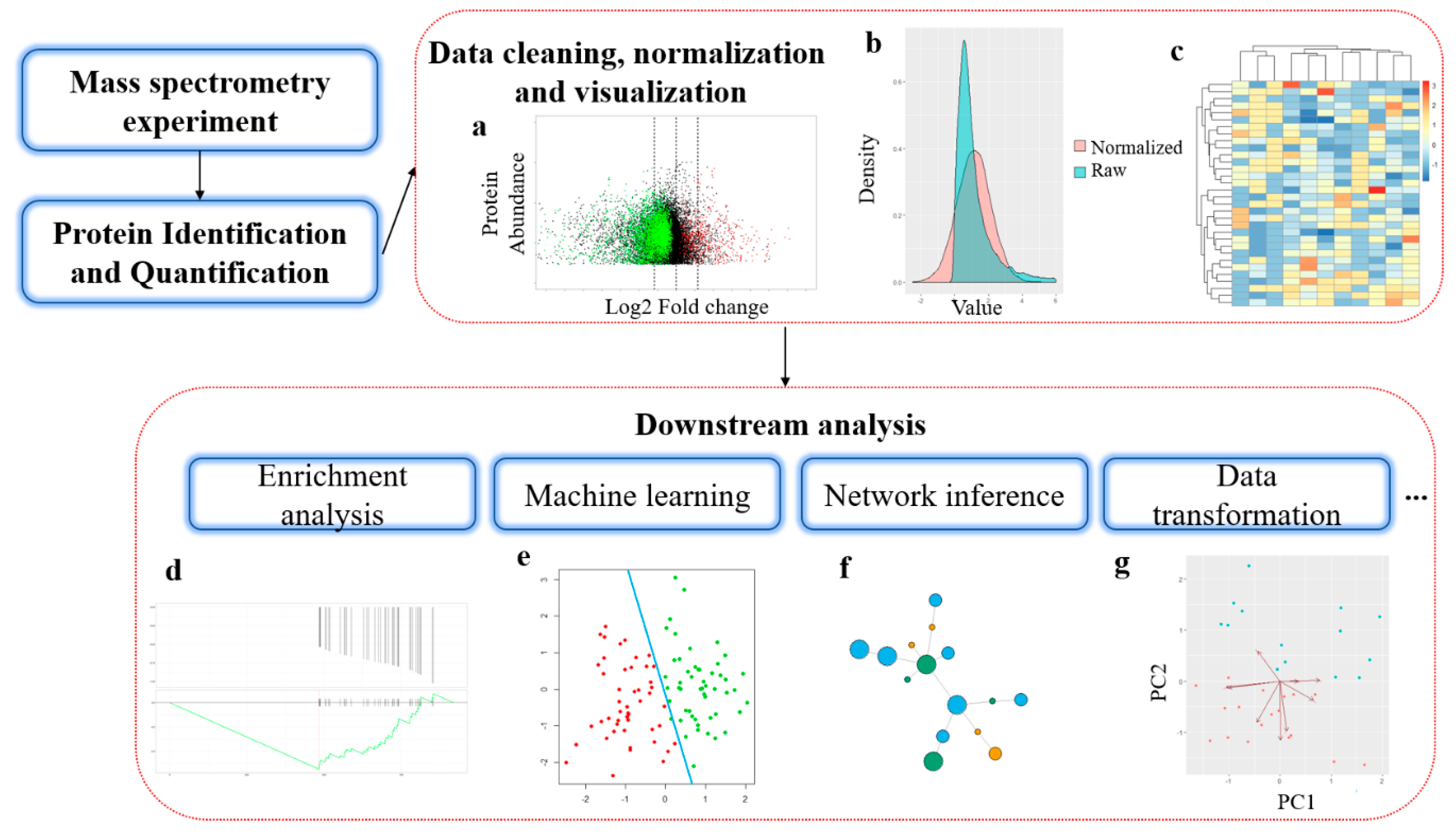



![Using genetic programming to predict and optimize protein function [PeerJ] Using genetic programming to predict and optimize protein function [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/pchem-24/1/fig-2-full.png)